Abstract
Background
Despite remarkable advances in our knowledge of epigenetically mediated transcriptional programming of cell differentiation in plants, little is known about chromatin topology and its functional implications in this process.
Results
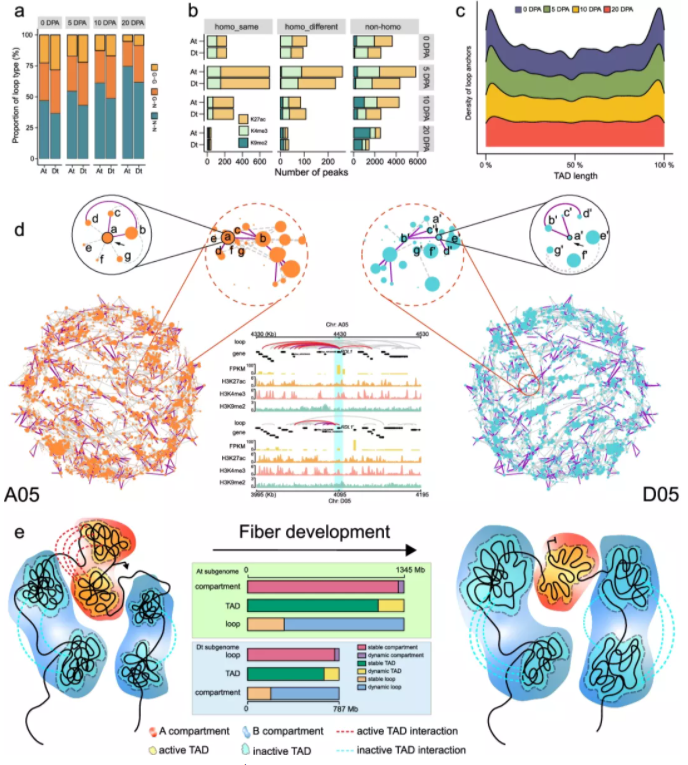
To interrogate its significance, we establish the dynamic three-dimensional (3D) genome architecture of the allotetraploid cotton fiber, representing a typical single cell undergoing staged development in plants. We show that the subgenome-relayed switching of the chromatin compartment from active to inactive is coupled with the silencing of developmentally repressed genes, pinpointing subgenome-coordinated contribution to fiber development. We identify 10,571 topologically associating domain-like (TAD-like) structures, of which 25.6% are specifically organized in different stages and 75.23% are subject to partition or fusion between two subgenomes. Notably, dissolution of intricate TAD-like structure cliques showing long-range interactions represents a prominent characteristic at the later developmental stage. Dynamic chromatin loops are found to mediate the rewiring of gene regulatory networks that exhibit a significant difference between the two subgenomes, implicating expression bias of homologous genes.
Conclusions
This study sheds light on the spatial-temporal asymmetric chromatin structures of two subgenomes in the cotton fiber and offers a new insight into the regulatory orchestration of cell differentiation in plants.
Full text:https://genomebiology.biomedcentral.com/articles/10.1186/s13059-022-02616-y